Introduction and Data Structures
Contents
# Install the necessary dependencies
import os
import sys
import numpy as np
import pandas as pd
!{sys.executable} -m pip install --quiet jupyterlab_myst ipython
5.4.1. Introduction and Data Structures#
Pandas is a fast, powerful, flexible and easy-to-use open-source data analysis and manipulation tool, built on top of the Python programming language.
5.4.1.1. Introducing Pandas objects#
In 3 sections, we’ll start with a quick, non-comprehensive overview of the fundamental data structures in Pandas to get you started. The fundamental behavior about data types, indexing, axis labeling, and alignment apply across all of the objects.
5.4.1.2. Overview#
In this section, we’ll introduce two data structure of pandas and the basic concept of data indexing and selection.
5.4.1.2.1. Series#
Series is a one-dimensional labeled array capable of holding any data type (integers, strings, floating point numbers, Python objects, etc.). The axis labels are collectively referred to as the index. The basic method to create a Series is to call:
s = pd.Series(data, index=index)
Here, data can be many different things:
a Python dict
an ndarray
a scalar value (like 5)
The passed index is a list of axis labels. Thus, this separates into a few cases depending on what the data is:
5.4.1.2.1.1. Create a Series#
5.4.1.2.1.1.1. From ndarray#
If data is an ndarray, index must be the same length as the data. If no index is passed, one will be created having values [0, ..., len(data) - 1].
s = pd.Series(np.random.randn(5), index=["a", "b", "c", "d", "e"])
s
a -0.648682
b -0.890614
c -1.669571
d 0.522419
e 0.075649
dtype: float64
s.index
Index(['a', 'b', 'c', 'd', 'e'], dtype='object')
pd.Series(np.random.randn(5))
0 -1.324419
1 -1.426718
2 1.169354
3 -0.688856
4 1.164765
dtype: float64
Note
Pandas supports non-unique index values. If an operation that does not support duplicate index values is attempted, an exception will be raised at that time.
5.4.1.2.1.1.2. From dict#
Series can be instantiated from dicts:
d = {"b": 1, "a": 0, "c": 2}
pd.Series(d)
b 1
a 0
c 2
dtype: int64
If an index is passed, the values in data corresponding to the labels in the index will be pulled out.
d = {"a": 0.0, "b": 1.0, "c": 2.0}
pd.Series(d)
a 0.0
b 1.0
c 2.0
dtype: float64
pd.Series(d, index=["b", "c", "d", "a"])
b 1.0
c 2.0
d NaN
a 0.0
dtype: float64
Note
NaN (not a number) is the standard missing data marker used in Pandas.
5.4.1.2.1.1.3. From scalar value#
If data is a scalar value, an index must be provided. The value will be repeated to match the length of index.
pd.Series(5.0, index=["a", "b", "c", "d", "e"])
a 5.0
b 5.0
c 5.0
d 5.0
e 5.0
dtype: float64
5.4.1.2.1.2. Series is ndarray-like#
Series acts very similarly to a ndarray and is a valid argument to most NumPy functions. However, operations such as slicing will also slice the index.
s[0]
-0.6486820579248709
s[:3]
a -0.648682
b -0.890614
c -1.669571
dtype: float64
s[s > s.median()]
d 0.522419
e 0.075649
dtype: float64
s[[4, 3, 1]]
e 0.075649
d 0.522419
b -0.890614
dtype: float64
np.exp(s)
a 0.522734
b 0.410404
c 0.188328
d 1.686102
e 1.078584
dtype: float64
Like a NumPy array, a Pandas Series has a single dtype.
s.dtype
dtype('float64')
If you need the actual array backing a Series, use Series.array.
s.array
<PandasArray>
[-0.6486820579248709, -0.890613503036024, -1.6695709617868482,
0.5224193821349435, 0.07564878277449659]
Length: 5, dtype: float64
While Series is ndarray-like, if you need an actual ndarray, then use Series.to_numpy().
s.to_numpy()
array([-0.64868206, -0.8906135 , -1.66957096, 0.52241938, 0.07564878])
Even if the Series is backed by an ExtensionArray, Series.to_numpy() will return a NumPy ndarray.
5.4.1.2.1.3. Series is dict-like#
A Series is also like a fixed-size dict in that you can get and set values by index label:
s["a"]
-0.6486820579248709
s["e"] = 12.0
s
a -0.648682
b -0.890614
c -1.669571
d 0.522419
e 12.000000
dtype: float64
"e" in s
True
"f" in s
False
If a label is not contained in the index, an exception is raised:
s["f"]
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/core/indexes/base.py:3803, in Index.get_loc(self, key, method, tolerance)
3802 try:
-> 3803 return self._engine.get_loc(casted_key)
3804 except KeyError as err:
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/_libs/index.pyx:138, in pandas._libs.index.IndexEngine.get_loc()
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/_libs/index.pyx:165, in pandas._libs.index.IndexEngine.get_loc()
File pandas/_libs/hashtable_class_helper.pxi:5745, in pandas._libs.hashtable.PyObjectHashTable.get_item()
File pandas/_libs/hashtable_class_helper.pxi:5753, in pandas._libs.hashtable.PyObjectHashTable.get_item()
KeyError: 'f'
The above exception was the direct cause of the following exception:
KeyError Traceback (most recent call last)
Cell In[25], line 1
----> 1 s["f"]
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/core/series.py:981, in Series.__getitem__(self, key)
978 return self._values[key]
980 elif key_is_scalar:
--> 981 return self._get_value(key)
983 if is_hashable(key):
984 # Otherwise index.get_value will raise InvalidIndexError
985 try:
986 # For labels that don't resolve as scalars like tuples and frozensets
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/core/series.py:1089, in Series._get_value(self, label, takeable)
1086 return self._values[label]
1088 # Similar to Index.get_value, but we do not fall back to positional
-> 1089 loc = self.index.get_loc(label)
1090 return self.index._get_values_for_loc(self, loc, label)
File /usr/share/miniconda/envs/open-machine-learning-jupyter-book/lib/python3.9/site-packages/pandas/core/indexes/base.py:3805, in Index.get_loc(self, key, method, tolerance)
3803 return self._engine.get_loc(casted_key)
3804 except KeyError as err:
-> 3805 raise KeyError(key) from err
3806 except TypeError:
3807 # If we have a listlike key, _check_indexing_error will raise
3808 # InvalidIndexError. Otherwise we fall through and re-raise
3809 # the TypeError.
3810 self._check_indexing_error(key)
KeyError: 'f'
Using the Series.get() method, a missing label will return None or specified default:
s.get("f")
s.get("f", np.nan)
nan
These labels can also be accessed by attribute.
5.4.1.2.1.4. Vectorized operations and label alignment with Series#
When working with raw NumPy arrays, looping through value-by-value is usually not necessary. The same is true when working with Series in Pandas. Series can also be passed into most NumPy methods expecting an ndarray.
s + s
a -1.297364
b -1.781227
c -3.339142
d 1.044839
e 24.000000
dtype: float64
s * 2
a -1.297364
b -1.781227
c -3.339142
d 1.044839
e 24.000000
dtype: float64
np.exp(s)
a 0.522734
b 0.410404
c 0.188328
d 1.686102
e 162754.791419
dtype: float64
A key difference between Series and ndarray is that operations between Series automatically align the data based on the label. Thus, you can write computations without giving consideration to whether the Series involved have the same labels.
s[1:] + s[:-1]
a NaN
b -1.781227
c -3.339142
d 1.044839
e NaN
dtype: float64
The result of an operation between unaligned Series will have the union of the indexes involved. If a label is not found in one Series or the other, the result will be marked as missing NaN. Being able to write code without doing any explicit data alignment grants immense freedom and flexibility in interactive data analysis and research. The integrated data alignment features of the Pandas data structures set Pandas apart from the majority of related tools for working with labeled data.
Note
In general, we chose to make the default result of operations between differently indexed objects yield the union of the indexes in order to avoid loss of information. Having an index label, though the data is missing, is typically important information as part of a computation. You of course have the option of dropping labels with missing data via the dropna function.
5.4.1.2.1.5. Name attribute#
Series also has a name attribute:
s = pd.Series(np.random.randn(5), name="something")
s
0 0.740679
1 -0.147724
2 0.204340
3 0.904797
4 0.225220
Name: something, dtype: float64
s.name
'something'
The Series name can be assigned automatically in many cases, in particular, when selecting a single column from a DataFrame, the name will be assigned the column label.
You can rename a Series with the pandas.Series.rename() method.
s2 = s.rename("different")
s2.name
'different'
Note that s and s2 refer to different objects.
5.4.1.2.2. DataFrame#
DataFrame is a 2-dimensional labeled data structure with columns of potentially different types. You can think of it like a spreadsheet or SQL table, or a dict of Series objects. It is generally the most commonly used Pandas object. Like Series, DataFrame accepts many different kinds of input:
Dict of 1D ndarrays, lists, dicts, or
Series2-D
numpy.ndarrayStructured or record ndarray
A
SeriesAnother
DataFrame
Along with the data, you can optionally pass index (row labels) and columns (column labels) arguments. If you pass an index and / or columns, you are guaranteeing the index and / or columns of the resulting DataFrame. Thus, a dict of Series plus a specific index will discard all data not matching up to the passed index.
If axis labels are not passed, they will be constructed from the input data based on common sense rules.
5.4.1.2.2.1. Create a Dataframe#
5.4.1.2.2.1.1. From dict of Series or dicts#
The resulting index will be the union of the indexes of the various Series. If there are any nested dicts, these will first be converted to Series. If no columns are passed, the columns will be the ordered list of dict keys.
d = {
"one": pd.Series([1.0, 2.0, 3.0], index=["a", "b", "c"]),
"two": pd.Series([1.0, 2.0, 3.0, 4.0], index=["a", "b", "c", "d"]),
}
df = pd.DataFrame(d)
df
| one | two | |
|---|---|---|
| a | 1.0 | 1.0 |
| b | 2.0 | 2.0 |
| c | 3.0 | 3.0 |
| d | NaN | 4.0 |
pd.DataFrame(d, index=["d", "b", "a"])
| one | two | |
|---|---|---|
| d | NaN | 4.0 |
| b | 2.0 | 2.0 |
| a | 1.0 | 1.0 |
pd.DataFrame(d, index=["d", "b", "a"], columns=["two", "three"])
| two | three | |
|---|---|---|
| d | 4.0 | NaN |
| b | 2.0 | NaN |
| a | 1.0 | NaN |
The row and column labels can be accessed respectively by accessing the index and columns attributes:
Note
When a particular set of columns is passed along with a dict of data, the passed columns override the keys in the dict.
df.index
Index(['a', 'b', 'c', 'd'], dtype='object')
df.columns
Index(['one', 'two'], dtype='object')
5.4.1.2.2.1.2. From dict of ndarrays / lists#
The ndarrays must all be the same length. If an index is passed, it must also be the same length as the arrays. If no index is passed, the result will be range(n), where n is the array length.
d = {"one": [1.0, 2.0, 3.0, 4.0], "two": [4.0, 3.0, 2.0, 1.0]}
pd.DataFrame(d)
| one | two | |
|---|---|---|
| 0 | 1.0 | 4.0 |
| 1 | 2.0 | 3.0 |
| 2 | 3.0 | 2.0 |
| 3 | 4.0 | 1.0 |
pd.DataFrame(d, index=["a", "b", "c", "d"])
| one | two | |
|---|---|---|
| a | 1.0 | 4.0 |
| b | 2.0 | 3.0 |
| c | 3.0 | 2.0 |
| d | 4.0 | 1.0 |
5.4.1.2.2.1.3. From structured or record array#
This case is handled identically to a dict of arrays.
data = np.zeros((2,), dtype=[("A", "i4"), ("B", "f4"), ("C", "a10")])
data[:] = [(1, 2.0, "Hello"), (2, 3.0, "World")]
pd.DataFrame(data)
| A | B | C | |
|---|---|---|---|
| 0 | 1 | 2.0 | b'Hello' |
| 1 | 2 | 3.0 | b'World' |
pd.DataFrame(data, index=["first", "second"])
| A | B | C | |
|---|---|---|---|
| first | 1 | 2.0 | b'Hello' |
| second | 2 | 3.0 | b'World' |
pd.DataFrame(data, columns=["C", "A", "B"])
| C | A | B | |
|---|---|---|---|
| 0 | b'Hello' | 1 | 2.0 |
| 1 | b'World' | 2 | 3.0 |
Note
DataFrame is not intended to work exactly like a 2-dimensional NumPy ndarray.
5.4.1.2.2.1.4. From a list of dicts#
data2 = [{"a": 1, "b": 2}, {"a": 5, "b": 10, "c": 20}]
pd.DataFrame(data2)
| a | b | c | |
|---|---|---|---|
| 0 | 1 | 2 | NaN |
| 1 | 5 | 10 | 20.0 |
pd.DataFrame(data2, index=["first", "second"])
| a | b | c | |
|---|---|---|---|
| first | 1 | 2 | NaN |
| second | 5 | 10 | 20.0 |
pd.DataFrame(data2, columns=["a", "b"])
| a | b | |
|---|---|---|
| 0 | 1 | 2 |
| 1 | 5 | 10 |
5.4.1.2.2.1.5. From a dict of tuples#
You can automatically create a MultiIndexed frame by passing a tuples dictionary.
pd.DataFrame(
{
("a", "b"): {("A", "B"): 1, ("A", "C"): 2},
("a", "a"): {("A", "C"): 3, ("A", "B"): 4},
("a", "c"): {("A", "B"): 5, ("A", "C"): 6},
("b", "a"): {("A", "C"): 7, ("A", "B"): 8},
("b", "b"): {("A", "D"): 9, ("A", "B"): 10},
}
)
| a | b | |||||
|---|---|---|---|---|---|---|
| b | a | c | a | b | ||
| A | B | 1.0 | 4.0 | 5.0 | 8.0 | 10.0 |
| C | 2.0 | 3.0 | 6.0 | 7.0 | NaN | |
| D | NaN | NaN | NaN | NaN | 9.0 | |
5.4.1.2.2.1.6. From a Series#
The result will be a DataFrame with the same index as the input Series, and with one column whose name is the original name of the Series (only if no other column name provided).
ser = pd.Series(range(3), index=list("abc"), name="ser")
pd.DataFrame(ser)
| ser | |
|---|---|
| a | 0 |
| b | 1 |
| c | 2 |
5.4.1.2.2.1.7. From a list of namedtuples#
The field names of the first namedtuple in the list determine the columns of the DataFrame. The remaining namedtuples (or tuples) are simply unpacked and their values are fed into the rows of the DataFrame. If any of those tuples is shorter than the first namedtuple then the later columns in the corresponding row are marked as missing values. If any are longer than the first namedtuple , a ValueError is raised.
from collections import namedtuple
Point = namedtuple("Point", "x y")
pd.DataFrame([Point(0, 0), Point(0, 3), (2, 3)])
| x | y | |
|---|---|---|
| 0 | 0 | 0 |
| 1 | 0 | 3 |
| 2 | 2 | 3 |
Point3D = namedtuple("Point3D", "x y z")
pd.DataFrame([Point3D(0, 0, 0), Point3D(0, 3, 5), Point(2, 3)])
| x | y | z | |
|---|---|---|---|
| 0 | 0 | 0 | 0.0 |
| 1 | 0 | 3 | 5.0 |
| 2 | 2 | 3 | NaN |
5.4.1.2.2.1.8. From a list of dataclasses#
Data Classes as introduced in PEP557, can be passed into the DataFrame constructor. Passing a list of dataclasses is equivalent to passing a list of dictionaries.
Please be aware, that all values in the list should be dataclasses, mixing types in the list would result in a TypeError.
from dataclasses import make_dataclass
Point = make_dataclass("Point", [("x", int), ("y", int)])
pd.DataFrame([Point(0, 0), Point(0, 3), Point(2, 3)])
| x | y | |
|---|---|---|
| 0 | 0 | 0 |
| 1 | 0 | 3 |
| 2 | 2 | 3 |
5.4.1.2.2.2. Column selection, addition, deletion#
You can treat a DataFrame semantically like a dict of like-indexed Series objects. Getting, setting, and deleting columns works with the same syntax as the analogous dict operations:
df
| one | two | |
|---|---|---|
| a | 1.0 | 1.0 |
| b | 2.0 | 2.0 |
| c | 3.0 | 3.0 |
| d | NaN | 4.0 |
df["one"]
a 1.0
b 2.0
c 3.0
d NaN
Name: one, dtype: float64
df["three"] = df["one"] * df["two"]
df["flag"] = df["one"] > 2
df
| one | two | three | flag | |
|---|---|---|---|---|
| a | 1.0 | 1.0 | 1.0 | False |
| b | 2.0 | 2.0 | 4.0 | False |
| c | 3.0 | 3.0 | 9.0 | True |
| d | NaN | 4.0 | NaN | False |
Columns can be deleted or popped like with a dict:
del df["two"]
three = df.pop("three")
df
| one | flag | |
|---|---|---|
| a | 1.0 | False |
| b | 2.0 | False |
| c | 3.0 | True |
| d | NaN | False |
When inserting a scalar value, it will naturally be propagated to fill the column:
df["foo"] = "bar"
df
| one | flag | foo | |
|---|---|---|---|
| a | 1.0 | False | bar |
| b | 2.0 | False | bar |
| c | 3.0 | True | bar |
| d | NaN | False | bar |
When inserting a Series that does not have the same index as the DataFrame, it will be conformed to the DataFrame’s index:
df["one_trunc"] = df["one"][:2]
from IPython.display import HTML
display(
HTML(
"""
<link rel="stylesheet" href="https://ocademy-ai.github.io/machine-learning/_static/style.css">
<div class='full-width docutils' >
<div class="admonition note pandastutor" name="html-admonition" style="margin-right:20%">
<p class="admonition-title pandastutor">Let's visualize it! 🎥</p>
<div class="pandastutor inner" style="height:730px;">
<iframe frameborder="0" scrolling="no" src="https://pandastutor.com/vis.html#code=import%20pandas%20as%20pd%0Aimport%20io%0Aimport%20numpy%20as%20np%0Ad%20%3D%20%7B%0A%20%20%20%20%22one%22%3A%20pd.Series%28%5B1.0,%202.0,%203.0%5D,%20index%3D%5B%22a%22,%20%22b%22,%20%22c%22%5D%29,%0A%20%20%20%20%22two%22%3A%20pd.Series%28%5B1.0,%202.0,%203.0,%204.0%5D,%20index%3D%5B%22a%22,%20%22b%22,%20%22c%22,%20%22d%22%5D%29,%0A%7D%0Adf%20%3D%20pd.DataFrame%28d%29%0Adf%5B%22flag%22%5D%20%3D%20df%5B%22one%22%5D%20%3E%202%0Adel%20df%5B%22two%22%5D%0Adf%5B%22foo%22%5D%20%3D%20%22bar%22%0Adf%5B%22one_trunc%22%5D%20%3Ddf.insert%281,%20%22bar%22,%20df%5B%22one%22%5D%29%0Adf.loc%5B%22b%22%5D&d=2023-09-19&lang=py&v=v1
"> </iframe>
</div>
</div>
</div>
"""
)
)
Let's visualize it! 🎥
df
| one | flag | foo | one_trunc | |
|---|---|---|---|---|
| a | 1.0 | False | bar | 1.0 |
| b | 2.0 | False | bar | 2.0 |
| c | 3.0 | True | bar | NaN |
| d | NaN | False | bar | NaN |
You can insert raw ndarrays but their length must match the length of the DataFrame’s index.
By default, columns get inserted at the end. DataFrame.insert() inserts at a particular location in the columns:
df.insert(1, "bar", df["one"])
df
| one | bar | flag | foo | one_trunc | |
|---|---|---|---|---|---|
| a | 1.0 | 1.0 | False | bar | 1.0 |
| b | 2.0 | 2.0 | False | bar | 2.0 |
| c | 3.0 | 3.0 | True | bar | NaN |
| d | NaN | NaN | False | bar | NaN |
5.4.1.2.2.3. Assigning new columns in method chains#
DataFrame has an assign() method that allows you to easily create new columns that are potentially derived from existing columns.
iris = pd.read_csv("https://static-1300131294.cos.ap-shanghai.myqcloud.com/data/data-science/working-with-data/pandas/iris.csv")
iris.head()
| SepalLength | SepalWidth | PetalLength | PetalWidth | Name | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa |
iris.assign(sepal_ratio=iris["SepalWidth"] / iris["SepalLength"]).head()
| SepalLength | SepalWidth | PetalLength | PetalWidth | Name | sepal_ratio | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa | 0.686275 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa | 0.612245 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa | 0.680851 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | 0.673913 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | 0.720000 |
In the example above, we inserted a precomputed value. We can also pass in a function of one argument to be evaluated on the DataFrame being assigned to.
iris.assign(sepal_ratio=lambda x: (x["SepalWidth"] / x["SepalLength"])).head()
| SepalLength | SepalWidth | PetalLength | PetalWidth | Name | sepal_ratio | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | Iris-setosa | 0.686275 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | Iris-setosa | 0.612245 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | Iris-setosa | 0.680851 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | Iris-setosa | 0.673913 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | Iris-setosa | 0.720000 |
assign() always returns a copy of the data, leaving the original DataFrame untouched.
Passing a callable, as opposed to an actual value to be inserted, is useful when you don’t have a reference to the DataFrame at hand. This is common when using assign() in a chain of operations. For example, we can limit the DataFrame to just those observations with a Sepal Length greater than 5, calculate the ratio, and plot:
(
iris.query("SepalLength > 5")
.assign(
SepalRatio=lambda x: x.SepalWidth / x.SepalLength,
PetalRatio=lambda x: x.PetalWidth / x.PetalLength,
)
.plot(kind="scatter", x="SepalRatio", y="PetalRatio")
)
<AxesSubplot: xlabel='SepalRatio', ylabel='PetalRatio'>
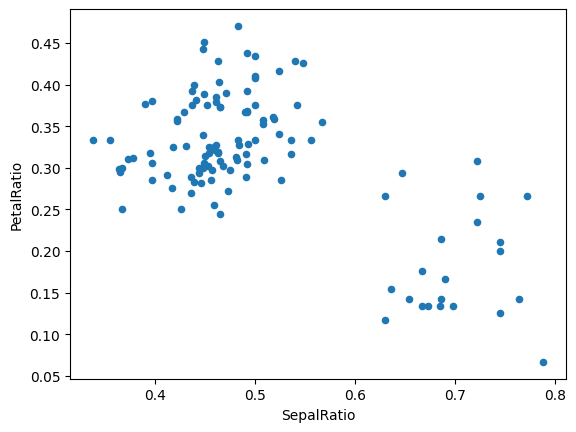
Since a function is passed in, the function is computed on the DataFrame being assigned to. Importantly, this is the DataFrame that’s been filtered to those rows with sepal length greater than 5. The filtering happens first, and then the ratio calculations. This is an example where we didn’t have a reference to the filtered DataFrame available.
The function signature for assign() is simply **kwargs. The keys are the column names for the new fields, and the values are either a value to be inserted (for example, a Series or NumPy array), or a function of one argument to be called on the DataFrame. A copy of the original DataFrame is returned, with the new values inserted.
The order of **kwargs is preserved. This allows for dependent assignment, where an expression later in **kwargs can refer to a column created earlier in the same assign().
dfa = pd.DataFrame({"A": [1, 2, 3], "B": [4, 5, 6]})
dfa.assign(C=lambda x: x["A"] + x["B"], D=lambda x: x["A"] + x["C"])
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 1 | 4 | 5 | 6 |
| 1 | 2 | 5 | 7 | 9 |
| 2 | 3 | 6 | 9 | 12 |
In the second expression, x['C'] will refer to the newly created column, that’s equal to dfa['A'] + dfa['B'].
5.4.1.2.2.4. Indexing / selection#
The basics of indexing are as follows:
Operation |
Syntax |
Result |
|---|---|---|
Select column |
|
Series |
Select row by label |
|
Series |
Select row by integer location |
|
Series |
Slice rows |
|
DataFrame |
Select rows by boolean vector |
|
DataFrame |
Row selection, for example, returns a Series whose index is the columns of the DataFrame:
df.loc["b"]
one 2.0
bar 2.0
flag False
foo bar
one_trunc 2.0
Name: b, dtype: object
from IPython.display import HTML
display(
HTML(
"""
<link rel="stylesheet" href="https://ocademy-ai.github.io/machine-learning/_static/style.css">
<div class='full-width docutils' >
<div class="admonition note pandastutor" name="html-admonition" style="margin-right:20%">
<p class="admonition-title pandastutor">Let's visualize it! 🎥</p>
<div class="pandastutor inner" style="height:730px;">
<iframe frameborder="0" scrolling="no" src="https://pandastutor.com/vis.html#code=import%20pandas%20as%20pd%0Aimport%20io%0Aimport%20numpy%20as%20np%0Ad%20%3D%20%7B%0A%20%20%20%20%22one%22%3A%20pd.Series%28%5B1.0,%202.0,%203.0%5D,%20index%3D%5B%22a%22,%20%22b%22,%20%22c%22%5D%29,%0A%20%20%20%20%22two%22%3A%20pd.Series%28%5B1.0,%202.0,%203.0,%204.0%5D,%20index%3D%5B%22a%22,%20%22b%22,%20%22c%22,%20%22d%22%5D%29,%0A%7D%0Adf%20%3D%20pd.DataFrame%28d%29%0Adf%5B%22flag%22%5D%20%3D%20df%5B%22one%22%5D%20%3E%202%0Adel%20df%5B%22two%22%5D%0Adf%5B%22foo%22%5D%20%3D%20%22bar%22%0Adf%5B%22one_trunc%22%5D%20%3Ddf.insert%281,%20%22bar%22,%20df%5B%22one%22%5D%29%0Adf.loc%5B%22b%22%5D&d=2023-09-19&lang=py&v=v1"> </iframe>
</div>
</div>
</div>
"""
)
)
Let's visualize it! 🎥
df.iloc[2]
one 3.0
bar 3.0
flag True
foo bar
one_trunc NaN
Name: c, dtype: object
5.4.1.2.2.5. Data alignment and arithmetic#
Data alignment between DataFrame objects automatically aligns on both the columns and the index (row labels)**. Again, the resulting object will have the union of the column and row labels.
df = pd.DataFrame(np.random.randn(10, 4), columns=["A", "B", "C", "D"])
df2 = pd.DataFrame(np.random.randn(7, 3), columns=["A", "B", "C"])
df + df2
| A | B | C | D | |
|---|---|---|---|---|
| 0 | -1.065518 | 1.194950 | 0.030608 | NaN |
| 1 | 1.837983 | 0.316330 | -2.104771 | NaN |
| 2 | -1.381854 | 1.362421 | 1.120580 | NaN |
| 3 | 0.716434 | -0.667963 | -2.372925 | NaN |
| 4 | 1.813942 | -2.189745 | -1.778060 | NaN |
| 5 | -0.680451 | 0.198311 | -1.035426 | NaN |
| 6 | 0.070760 | 2.201920 | 1.783717 | NaN |
| 7 | NaN | NaN | NaN | NaN |
| 8 | NaN | NaN | NaN | NaN |
| 9 | NaN | NaN | NaN | NaN |
When doing an operation between DataFrame and Series, the default behavior is to align the Series index on the DataFrame columns, thus broadcasting row-wise. For example:
df - df.iloc[0]
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 1 | -0.153982 | 0.106456 | -0.982637 | -0.719556 |
| 2 | -2.575348 | 0.616172 | 0.576169 | -0.021143 |
| 3 | -0.806753 | -0.229733 | -1.968249 | -0.490483 |
| 4 | 0.546768 | 0.125705 | -2.357085 | -0.210174 |
| 5 | 0.595272 | 0.003298 | -1.887417 | 1.366075 |
| 6 | -0.927779 | 1.775162 | 0.936908 | 1.671263 |
| 7 | -1.156538 | 0.646578 | -0.663057 | -0.767951 |
| 8 | -1.152485 | 0.072190 | -2.148250 | -0.339899 |
| 9 | -0.101024 | -1.220136 | -0.606941 | 0.116618 |
Arithmetic operations with scalars operate element-wise:
df * 5 + 2
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 4.212800 | 2.056525 | 3.468874 | 2.697986 |
| 1 | 3.442890 | 2.588806 | -1.444312 | -0.899793 |
| 2 | -8.663939 | 5.137387 | 6.349719 | 2.592272 |
| 3 | 0.179037 | 0.907857 | -6.372371 | 0.245569 |
| 4 | 6.946641 | 2.685050 | -8.316553 | 1.647115 |
| 5 | 7.189161 | 2.073015 | -5.968211 | 9.528360 |
| 6 | -0.426094 | 10.932335 | 8.153416 | 11.054301 |
| 7 | -1.569888 | 5.289417 | 0.153589 | -1.141768 |
| 8 | -1.549626 | 2.417474 | -7.272374 | 0.998492 |
| 9 | 3.707681 | -4.044153 | 0.434168 | 3.281075 |
1 / df
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 2.259580 | 88.456943 | 3.403968 | 7.163469 |
| 1 | 3.465268 | 8.491757 | -1.451669 | -1.724261 |
| 2 | -0.468870 | 1.593683 | 1.149500 | 8.442063 |
| 3 | -2.745800 | -4.578157 | -0.597202 | -2.849926 |
| 4 | 1.010787 | 7.298739 | -0.484658 | -14.168931 |
| 5 | 0.963547 | 68.479290 | -0.627493 | 0.664155 |
| 6 | -2.060926 | 0.559764 | 0.812557 | 0.552224 |
| 7 | -1.400604 | 1.520026 | -2.707956 | -1.591461 |
| 8 | -1.408599 | 11.976801 | -0.539236 | -4.992471 |
| 9 | 2.927948 | -0.827246 | -3.193191 | 3.902973 |
df ** 4
| A | B | C | D | |
|---|---|---|---|---|
| 0 | 0.038361 | 1.633324e-08 | 0.007448 | 0.000380 |
| 1 | 0.006935 | 1.923135e-04 | 0.225180 | 0.113133 |
| 2 | 20.691434 | 1.550216e-01 | 0.572750 | 0.000197 |
| 3 | 0.017592 | 2.276341e-03 | 7.861654 | 0.015159 |
| 4 | 0.957991 | 3.523778e-04 | 18.124185 | 0.000025 |
| 5 | 1.160135 | 4.547398e-08 | 6.450052 | 5.139508 |
| 6 | 0.055431 | 1.018544e+01 | 2.293957 | 10.753247 |
| 7 | 0.259859 | 1.873250e-01 | 0.018597 | 0.155889 |
| 8 | 0.254010 | 4.860005e-05 | 11.827249 | 0.001610 |
| 9 | 0.013607 | 2.135315e+00 | 0.009618 | 0.004309 |
Boolean operators operate element-wise as well:
df1 = pd.DataFrame({"a": [1, 0, 1], "b": [0, 1, 1]}, dtype=bool)
df2 = pd.DataFrame({"a": [0, 1, 1], "b": [1, 1, 0]}, dtype=bool)
df1 & df2
| a | b | |
|---|---|---|
| 0 | False | False |
| 1 | False | True |
| 2 | True | False |
df1 | df2
| a | b | |
|---|---|---|
| 0 | True | True |
| 1 | True | True |
| 2 | True | True |
df1 ^ df2
| a | b | |
|---|---|---|
| 0 | True | True |
| 1 | True | False |
| 2 | False | True |
-df1
| a | b | |
|---|---|---|
| 0 | False | True |
| 1 | True | False |
| 2 | False | False |
5.4.1.2.2.6. Transposing#
To transpose, access the T attribute or DataFrame.transpose(), similar to an ndarray:
df[:5].T
| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| A | 0.442560 | 0.288578 | -2.132788 | -0.364193 | 0.989328 |
| B | 0.011305 | 0.117761 | 0.627477 | -0.218429 | 0.137010 |
| C | 0.293775 | -0.688862 | 0.869944 | -1.674474 | -2.063311 |
| D | 0.139597 | -0.579959 | 0.118454 | -0.350886 | -0.070577 |
5.4.1.3. Data indexing and selection#
The axis labeling information in Pandas objects serves many purposes:
Identifies data (i.e. provides metadata) using known indicators, important for analysis, visualization, and interactive console display.
Enables automatic and explicit data alignment.
Allows intuitive getting and setting of subsets of the data set.
In this section, we will focus on the final point: namely, how to slice, dice, and generally get and set subsets of Pandas objects. The primary focus will be on Series and DataFrame as they have received more development attention in this area.
Note
The Python and NumPy indexing operators [] and attribute operator . provide quick and easy access to Pandas data structures across a wide range of use cases. This makes interactive work intuitive, as there’s little new to learn if you already know how to deal with Python dictionaries and NumPy arrays. However, since the type of the data to be accessed isn’t known in advance, directly using standard operators has some optimization limits. For production code, we recommended that you take advantage of the optimized Pandas data access methods exposed in this chapter.
Whether a copy or a reference is returned for a setting operation, may depend on the context. This is sometimes called chained assignment and should be avoided.
5.4.1.3.1. Different choices for indexing#
Object selection has had a number of user-requested additions in order to support more explicit location-based indexing. Pandas now supports three types of multi-axis indexing.
.locis primarily label based, but may also be used with a boolean array..locwill raiseKeyErrorwhen the items are not found. Allowed inputs are:A single label, e.g.
5or'a'(Note that5is interpreted as a label of the index. This use is not an integer position along the index.).A list or array of labels
['a', 'b', 'c'].A slice object with labels
'a':'f'(Note that contrary to usual Python slices, both the start and the stop are included, when present in the index!)A boolean array (any
NAvalues will be treated asFalse).A
callablefunction with one argument (the calling Series or DataFrame) and that returns valid output for indexing (one of the above).
.ilocis primarily integer position based (from0tolength-1of the axis), but may also be used with a boolean array..ilocwill raiseIndexErrorif a requested indexer is out-of-bounds, except slice indexers which allow out-of-bounds indexing. (this conforms with Python/NumPy slice semantics). Allowed inputs are:An integer e.g.
5.A list or array of integers
[4, 3, 0].A slice object with ints
1:7.A boolean array (any
NAvalues will be treated asFalse).A
callablefunction with one argument (the calling Series or DataFrame) that returns valid output for indexing (one of the above).
.loc,.iloc, and also[]indexing can accept acallableas indexer.
Getting values from an object with multi-axes selection uses the following notation (using .loc as an example, but the following applies to .iloc as well). Any of the axes accessors may be the null slice :. Axes left out of the specification are assumed to be :, e.g. p.loc['a'] is equivalent to p.loc['a', :].
Object Type |
Indexers |
|---|---|
Series |
|
DataFrame |
|
5.4.1.3.2. Basics#
As mentioned when introducing the data structures in the last section, the primary function of indexing with [] (a.k.a. __getitem__ for those familiar with implementing class behavior in Python) is selecting out lower-dimensional slices. The following table shows return type values when indexing Pandas objects with []:
Object Type |
Selection |
Return Value Type |
|---|---|---|
Series |
|
scalar value |
DataFrame |
|
|
Here we construct a simple time series data set to use for illustrating the indexing functionality:
dates = pd.date_range('1/1/2000', periods=8)
df = pd.DataFrame(np.random.randn(8, 4),
index=dates, columns=['A', 'B', 'C', 'D'])
df
| A | B | C | D | |
|---|---|---|---|---|
| 2000-01-01 | 0.711418 | 0.603204 | -0.417841 | 1.391724 |
| 2000-01-02 | -0.744106 | 0.862873 | 1.216255 | -1.282529 |
| 2000-01-03 | -1.373171 | 1.581176 | 0.828784 | -2.048825 |
| 2000-01-04 | -0.588767 | -2.798199 | -0.018800 | 0.130053 |
| 2000-01-05 | 1.251814 | 0.068020 | 0.373987 | -0.041853 |
| 2000-01-06 | -0.377347 | 0.654981 | 0.059024 | 0.507564 |
| 2000-01-07 | 0.111746 | 0.016025 | -1.023215 | -0.677410 |
| 2000-01-08 | 1.409014 | 0.481835 | -0.756125 | 2.236706 |
Note
None of the indexing functionality is time series specific unless specifically stated.
Thus, as per above, we have the most basic indexing using []:
s = df['A']
s[dates[5]]
-0.3773466384496966
You can pass a list of columns to [] to select columns in that order. If a column is not contained in the DataFrame, an exception will be raised. Multiple columns can also be set in this manner:
df
| A | B | C | D | |
|---|---|---|---|---|
| 2000-01-01 | 0.711418 | 0.603204 | -0.417841 | 1.391724 |
| 2000-01-02 | -0.744106 | 0.862873 | 1.216255 | -1.282529 |
| 2000-01-03 | -1.373171 | 1.581176 | 0.828784 | -2.048825 |
| 2000-01-04 | -0.588767 | -2.798199 | -0.018800 | 0.130053 |
| 2000-01-05 | 1.251814 | 0.068020 | 0.373987 | -0.041853 |
| 2000-01-06 | -0.377347 | 0.654981 | 0.059024 | 0.507564 |
| 2000-01-07 | 0.111746 | 0.016025 | -1.023215 | -0.677410 |
| 2000-01-08 | 1.409014 | 0.481835 | -0.756125 | 2.236706 |
df[['B', 'A']] = df[['A', 'B']]
df
| A | B | C | D | |
|---|---|---|---|---|
| 2000-01-01 | 0.603204 | 0.711418 | -0.417841 | 1.391724 |
| 2000-01-02 | 0.862873 | -0.744106 | 1.216255 | -1.282529 |
| 2000-01-03 | 1.581176 | -1.373171 | 0.828784 | -2.048825 |
| 2000-01-04 | -2.798199 | -0.588767 | -0.018800 | 0.130053 |
| 2000-01-05 | 0.068020 | 1.251814 | 0.373987 | -0.041853 |
| 2000-01-06 | 0.654981 | -0.377347 | 0.059024 | 0.507564 |
| 2000-01-07 | 0.016025 | 0.111746 | -1.023215 | -0.677410 |
| 2000-01-08 | 0.481835 | 1.409014 | -0.756125 | 2.236706 |
You may find this useful for applying a transform (in-place) to a subset of the columns.
Pandas aligns all AXES when setting Series and DataFrame from .loc, and .iloc.
This will not modify df because the column alignment is before value assignment.
df[['A', 'B']]
| A | B | |
|---|---|---|
| 2000-01-01 | 0.603204 | 0.711418 |
| 2000-01-02 | 0.862873 | -0.744106 |
| 2000-01-03 | 1.581176 | -1.373171 |
| 2000-01-04 | -2.798199 | -0.588767 |
| 2000-01-05 | 0.068020 | 1.251814 |
| 2000-01-06 | 0.654981 | -0.377347 |
| 2000-01-07 | 0.016025 | 0.111746 |
| 2000-01-08 | 0.481835 | 1.409014 |
df.loc[:, ['B', 'A']] = df[['A', 'B']]
df[['A', 'B']]
| A | B | |
|---|---|---|
| 2000-01-01 | 0.603204 | 0.711418 |
| 2000-01-02 | 0.862873 | -0.744106 |
| 2000-01-03 | 1.581176 | -1.373171 |
| 2000-01-04 | -2.798199 | -0.588767 |
| 2000-01-05 | 0.068020 | 1.251814 |
| 2000-01-06 | 0.654981 | -0.377347 |
| 2000-01-07 | 0.016025 | 0.111746 |
| 2000-01-08 | 0.481835 | 1.409014 |
The correct way to swap column values is by using raw values:
df.loc[:, ['B', 'A']] = df[['A', 'B']].to_numpy()
df[['A', 'B']]
| A | B | |
|---|---|---|
| 2000-01-01 | 0.711418 | 0.603204 |
| 2000-01-02 | -0.744106 | 0.862873 |
| 2000-01-03 | -1.373171 | 1.581176 |
| 2000-01-04 | -0.588767 | -2.798199 |
| 2000-01-05 | 1.251814 | 0.068020 |
| 2000-01-06 | -0.377347 | 0.654981 |
| 2000-01-07 | 0.111746 | 0.016025 |
| 2000-01-08 | 1.409014 | 0.481835 |
5.4.1.3.3. Attribute access#
You may access an index on a Series or column on a DataFrame directly as an attribute:
sa = pd.Series([1, 2, 3], index=list('abc'))
dfa = df.copy()
sa.b
2
dfa.A
2000-01-01 0.711418
2000-01-02 -0.744106
2000-01-03 -1.373171
2000-01-04 -0.588767
2000-01-05 1.251814
2000-01-06 -0.377347
2000-01-07 0.111746
2000-01-08 1.409014
Freq: D, Name: A, dtype: float64
sa.a = 5
sa
a 5
b 2
c 3
dtype: int64
dfa.A = list(range(len(dfa.index))) # ok if A already exists
dfa
| A | B | C | D | |
|---|---|---|---|---|
| 2000-01-01 | 0 | 0.603204 | -0.417841 | 1.391724 |
| 2000-01-02 | 1 | 0.862873 | 1.216255 | -1.282529 |
| 2000-01-03 | 2 | 1.581176 | 0.828784 | -2.048825 |
| 2000-01-04 | 3 | -2.798199 | -0.018800 | 0.130053 |
| 2000-01-05 | 4 | 0.068020 | 0.373987 | -0.041853 |
| 2000-01-06 | 5 | 0.654981 | 0.059024 | 0.507564 |
| 2000-01-07 | 6 | 0.016025 | -1.023215 | -0.677410 |
| 2000-01-08 | 7 | 0.481835 | -0.756125 | 2.236706 |
dfa['A'] = list(range(len(dfa.index))) # use this form to create a new column
dfa
| A | B | C | D | |
|---|---|---|---|---|
| 2000-01-01 | 0 | 0.603204 | -0.417841 | 1.391724 |
| 2000-01-02 | 1 | 0.862873 | 1.216255 | -1.282529 |
| 2000-01-03 | 2 | 1.581176 | 0.828784 | -2.048825 |
| 2000-01-04 | 3 | -2.798199 | -0.018800 | 0.130053 |
| 2000-01-05 | 4 | 0.068020 | 0.373987 | -0.041853 |
| 2000-01-06 | 5 | 0.654981 | 0.059024 | 0.507564 |
| 2000-01-07 | 6 | 0.016025 | -1.023215 | -0.677410 |
| 2000-01-08 | 7 | 0.481835 | -0.756125 | 2.236706 |
You can use this access only if the index element is a valid Python identifier, e.g. s.1 is not allowed. See here for an explanation of valid identifiers.
The attribute will not be available if it conflicts with an existing method name, e.g. s.min is not allowed, but s[‘min’] is possible.
Similarly, the attribute will not be available if it conflicts with any of the following list: index, major_axis, minor_axis, items.
In any of these cases, standard indexing will still work, e.g. s[‘1’], s[‘min’], and s[‘index’] will access the corresponding element or column.
If you are using the IPython environment, you may also use tab-completion to see these accessible attributes.
You can also assign a dict to a row of a DataFrame:
x = pd.DataFrame({'x': [1, 2, 3], 'y': [3, 4, 5]})
x.iloc[1] = {'x': 9, 'y': 99}
x
| x | y | |
|---|---|---|
| 0 | 1 | 3 |
| 1 | 9 | 99 |
| 2 | 3 | 5 |
You can use attribute access to modify an existing element of a Series or column of a DataFrame, but be careful; if you try to use attribute access to create a new column, it creates a new attribute rather than a new column. In 0.21.0 and later, this will raise a UserWarning:
df = pd.DataFrame({'one': [1., 2., 3.]})
df.two = [4, 5, 6]
/tmp/ipykernel_2921/269534380.py:2: UserWarning: Pandas doesn't allow columns to be created via a new attribute name - see https://pandas.pydata.org/pandas-docs/stable/indexing.html#attribute-access
df.two = [4, 5, 6]
df
| one | |
|---|---|
| 0 | 1.0 |
| 1 | 2.0 |
| 2 | 3.0 |
5.4.1.3.4. Slicing ranges#
For now, we explain the semantics of slicing using the [] operator.
With Series, the syntax works exactly as with an ndarray, returning a slice of the values and the corresponding labels:
s
2000-01-01 0.711418
2000-01-02 -0.744106
2000-01-03 -1.373171
2000-01-04 -0.588767
2000-01-05 1.251814
2000-01-06 -0.377347
2000-01-07 0.111746
2000-01-08 1.409014
Freq: D, Name: A, dtype: float64
s[:5]
2000-01-01 0.711418
2000-01-02 -0.744106
2000-01-03 -1.373171
2000-01-04 -0.588767
2000-01-05 1.251814
Freq: D, Name: A, dtype: float64
s[::2]
2000-01-01 0.711418
2000-01-03 -1.373171
2000-01-05 1.251814
2000-01-07 0.111746
Freq: 2D, Name: A, dtype: float64
s[::-1]
2000-01-08 1.409014
2000-01-07 0.111746
2000-01-06 -0.377347
2000-01-05 1.251814
2000-01-04 -0.588767
2000-01-03 -1.373171
2000-01-02 -0.744106
2000-01-01 0.711418
Freq: -1D, Name: A, dtype: float64
Note that setting works as well:
s2 = s.copy()
s2[:5] = 0
s2
2000-01-01 0.000000
2000-01-02 0.000000
2000-01-03 0.000000
2000-01-04 0.000000
2000-01-05 0.000000
2000-01-06 -0.377347
2000-01-07 0.111746
2000-01-08 1.409014
Freq: D, Name: A, dtype: float64
With DataFrame, slicing inside of [] slices the rows. This is provided largely as a convenience since it is such a common operation.
df[:3]
| one | |
|---|---|
| 0 | 1.0 |
| 1 | 2.0 |
| 2 | 3.0 |
df[::-1]
| one | |
|---|---|
| 2 | 3.0 |
| 1 | 2.0 |
| 0 | 1.0 |
5.4.1.4. Acknowledgments#
Thanks for Pandas user guide. It contributes the majority of the content in this chapter.


