Support Vector Machines (SVM) - Intro and SVM for Regression
Contents
42.66. Support Vector Machines (SVM) - Intro and SVM for Regression#
Support Vector Machines are the type of supervised learning algorithms used for regression, classification and detecting outliers. SVMs are remarkably one of the powerful models in classical machine learning suited for handling complex and high dimensional datasets.
With SVM supporting different kernels (linear, polynomial, Radial Basis Function(rbf), and sigmoid), SVM can tackle different kinds of datasets, both linear and non linear.
While the maths behind the SVMs are beyond the scope of this notebook, here is the idea behind SVMs:
The way SVM works can be compared to a street with a boundary line. During SVM training, SMV draws the large margin or decision boundary between classes based on the importance of each training data point. The training data points that are inside the decision boundary are called support vectors and hence the name.
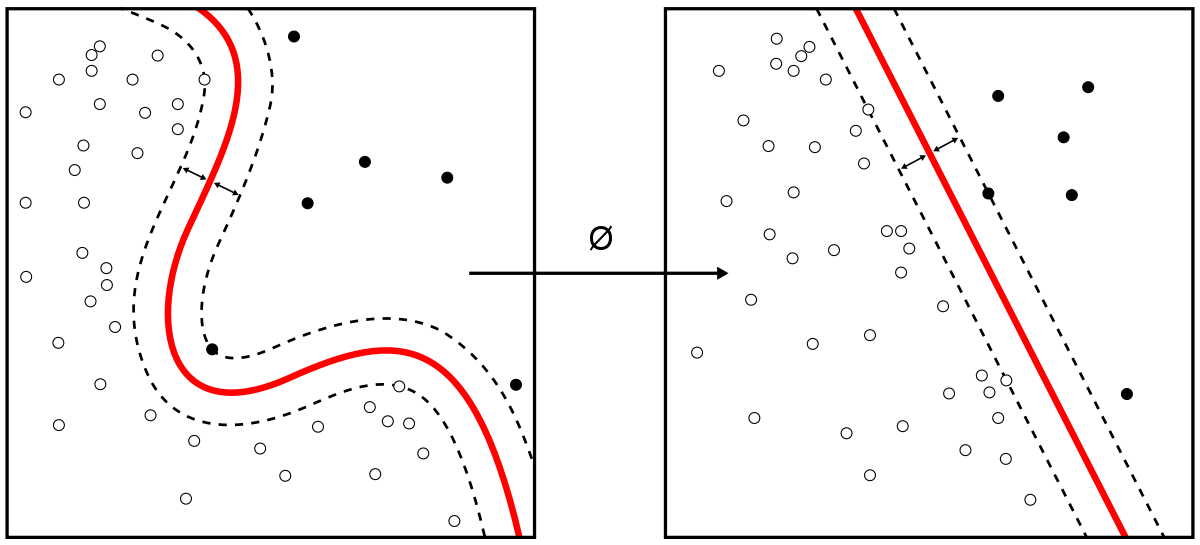
SMVs are widely used for classification. But to motivate that, let’s start with regression. For the purpose of examining how powerful this algorithm is, we will use the same dataset that we used in linear regression notebook, and hopefully the difference in performance will be notable.
42.66.1. Contents#
42.66.2. 1 - Imports#
import numpy as np
import pandas as pd
import seaborn as sns
import urllib.request
import sklearn
import matplotlib.pyplot as plt
%matplotlib inline
42.66.3. 2 - Loading the data#
cal_data = pd.read_csv("../../assets/data/housing.csv")
cal_data.head()
| longitude | latitude | housing_median_age | total_rooms | total_bedrooms | population | households | median_income | median_house_value | ocean_proximity | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | -122.23 | 37.88 | 41.0 | 880.0 | 129.0 | 322.0 | 126.0 | 8.3252 | 452600.0 | NEAR BAY |
| 1 | -122.22 | 37.86 | 21.0 | 7099.0 | 1106.0 | 2401.0 | 1138.0 | 8.3014 | 358500.0 | NEAR BAY |
| 2 | -122.24 | 37.85 | 52.0 | 1467.0 | 190.0 | 496.0 | 177.0 | 7.2574 | 352100.0 | NEAR BAY |
| 3 | -122.25 | 37.85 | 52.0 | 1274.0 | 235.0 | 558.0 | 219.0 | 5.6431 | 341300.0 | NEAR BAY |
| 4 | -122.25 | 37.85 | 52.0 | 1627.0 | 280.0 | 565.0 | 259.0 | 3.8462 | 342200.0 | NEAR BAY |
len(cal_data)
20640
42.66.4. 3 - Exploratory Analysis#
This is going to be a quick glance through the dataset. The full exploratory data analysis was done in the linear regression notebook[ADD LINK]. Before anything, let’s split the data into training and test set.
from sklearn.model_selection import train_test_split
train_data, test_data = train_test_split(cal_data, test_size=0.1,random_state=20)
print('The size of training data is: {} \nThe size of testing data is: {}'.format(len(train_data), len(test_data)))
The size of training data is: 18576
The size of testing data is: 2064
## Checking statistics
train_data.describe().transpose()
| count | mean | std | min | 25% | 50% | 75% | max | |
|---|---|---|---|---|---|---|---|---|
| longitude | 18576.0 | -119.567530 | 2.000581 | -124.3500 | -121.7900 | -118.4900 | -118.010000 | -114.4900 |
| latitude | 18576.0 | 35.630217 | 2.133260 | 32.5400 | 33.9300 | 34.2600 | 37.710000 | 41.9500 |
| housing_median_age | 18576.0 | 28.661068 | 12.604039 | 1.0000 | 18.0000 | 29.0000 | 37.000000 | 52.0000 |
| total_rooms | 18576.0 | 2631.567453 | 2169.467450 | 2.0000 | 1445.0000 | 2127.0000 | 3149.000000 | 39320.0000 |
| total_bedrooms | 18390.0 | 537.344698 | 417.672864 | 1.0000 | 295.0000 | 435.0000 | 648.000000 | 6445.0000 |
| population | 18576.0 | 1422.408376 | 1105.486111 | 3.0000 | 785.7500 | 1166.0000 | 1725.000000 | 28566.0000 |
| households | 18576.0 | 499.277078 | 379.473497 | 1.0000 | 279.0000 | 410.0000 | 606.000000 | 6082.0000 |
| median_income | 18576.0 | 3.870053 | 1.900225 | 0.4999 | 2.5643 | 3.5341 | 4.742725 | 15.0001 |
| median_house_value | 18576.0 | 206881.011305 | 115237.605962 | 14999.0000 | 120000.0000 | 179800.0000 | 264700.000000 | 500001.0000 |
## Checking missing values
train_data.isnull().sum()
longitude 0
latitude 0
housing_median_age 0
total_rooms 0
total_bedrooms 186
population 0
households 0
median_income 0
median_house_value 0
ocean_proximity 0
dtype: int64
# Checking Values in the Categorical Feature(s)
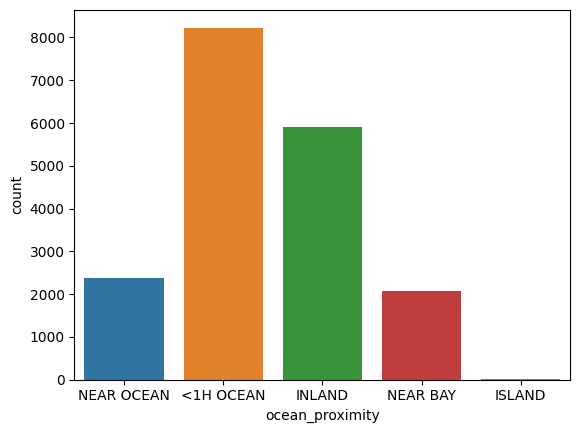
train_data['ocean_proximity'].value_counts()
<1H OCEAN 8231
INLAND 5896
NEAR OCEAN 2384
NEAR BAY 2061
ISLAND 4
Name: ocean_proximity, dtype: int64
sns.countplot(data=train_data, x='ocean_proximity')
<AxesSubplot: xlabel='ocean_proximity', ylabel='count'>
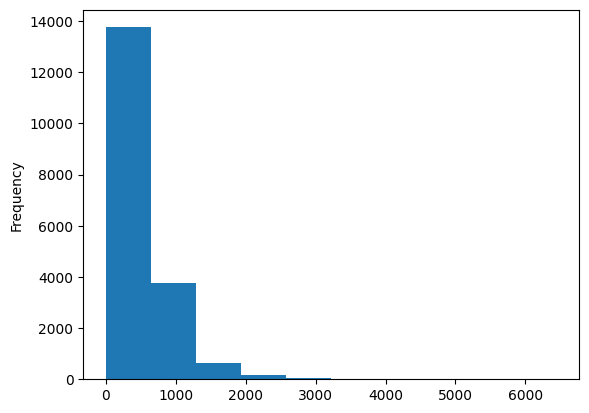
Total bedroom is the only feature having missing values. Here is its distribution.
train_data['total_bedrooms'].plot(kind='hist')
<AxesSubplot: ylabel='Frequency'>
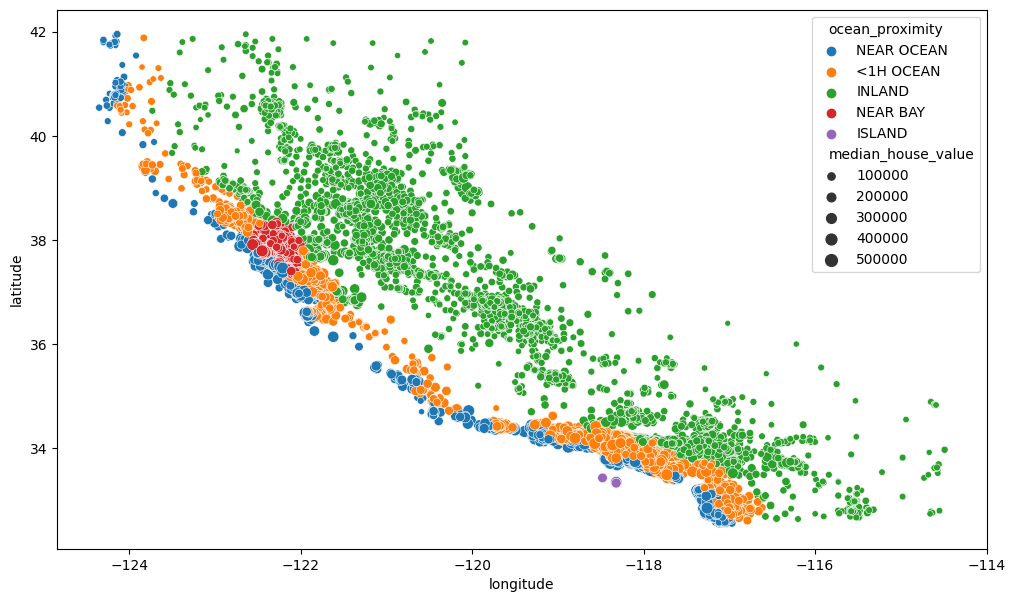
# Plotting geographical features
plt.figure(figsize=(12,7))
sns.scatterplot(data = train_data, x='longitude', y='latitude', hue='ocean_proximity',
size='median_house_value')
<AxesSubplot: xlabel='longitude', ylabel='latitude'>
42.66.5. 4 - Data Preprocessing#
To do:
Handle missing values
Encode categorical features
Scale the features
Everything into pipeline
# Getting training input data and labels
training_input_data = train_data.drop('median_house_value', axis=1)
training_labels = train_data['median_house_value']
# Numerical features
num_feats = training_input_data.drop('ocean_proximity', axis=1)
# Categorical features
cat_feats = training_input_data[['ocean_proximity']]
# Handle missing values
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import StandardScaler
num_pipe = Pipeline([
('imputer', SimpleImputer(strategy='mean')),
('scaler', StandardScaler())
])
num_preprocessed = num_pipe.fit_transform(num_feats)
# Pipeline to combine the numerical pipeline and also encode categorical features
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
# The transformer requires lists of features
num_list = list(num_feats)
cat_list = list(cat_feats)
final_pipe = ColumnTransformer([
('num', num_pipe, num_list),
('cat', OneHotEncoder(), cat_list)
])
training_data_preprocessed = final_pipe.fit_transform(training_input_data)
training_data_preprocessed
array([[ 0.67858615, -0.85796668, 0.97899282, ..., 0. ,
0. , 1. ],
[-0.93598814, 0.41242353, 0.18557502, ..., 0. ,
0. , 0. ],
[-1.45585107, 0.9187045 , 0.02689146, ..., 0. ,
0. , 1. ],
...,
[ 1.27342931, -1.32674535, 0.18557502, ..., 0. ,
0. , 0. ],
[ 0.64859406, -0.71733307, 0.97899282, ..., 0. ,
0. , 0. ],
[-1.44085502, 1.01246024, 1.37570172, ..., 0. ,
1. , 0. ]])
42.66.6. 5 - Training Support Vector Regressor#
In regression, instead of separating the classes with decision boundary like in classification, SVR fits the training data points on the boundary margin but keep them off.
We can implement it quite easily with Scikit-Learn.
from sklearn.svm import LinearSVR, SVR
lin_svr = LinearSVR()
lin_svr.fit(training_data_preprocessed, training_labels)
LinearSVR()In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
LinearSVR()
We can also use nonlinear SVM with a polynomial kernel function.
poly_svr = SVR(kernel='poly')
poly_svr.fit(training_data_preprocessed, training_labels)
SVR(kernel='poly')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
SVR(kernel='poly')
42.66.7. 6 - Evaluating Support Vector Regressor#
Let us evaluate the two models we created on the training set before evaluating on the test set. This is a good practice. Since finding a good model can take many iterations of improvements, it’s not advised to touch the test set until the model is good enough. Otherwise it would fail to make predictions on the new data.
As always, we evaluate regression models with mean squarred error, but the commonly used one is root mean squarred error.
from sklearn.metrics import mean_squared_error
predictions = lin_svr.predict(training_data_preprocessed)
mse = mean_squared_error(training_labels, predictions)
rmse = np.sqrt(mse)
rmse
215682.86713461558
That’s too high error. Let’s see how SVR with polynomial kernel will perform.
predictions = poly_svr.predict(training_data_preprocessed)
mse = mean_squared_error(training_labels, predictions)
rmse = np.sqrt(mse)
rmse
117513.38828582528
It did well than the former. For now, we can try to improve this with Randomized Search where we can find the best parameters. Let’s do that!
42.66.8. 7 - Improving Support Vector Regressor#
Let use Randomized Search to improve the SVR model. Few notes about the parameters:
Gamma(y): This is a regularization hyperparameter. When gamma is small, the model can underfit. It is too high, model can overfit.
C: It’s same as gamma. It is a regularization hyperparameter. When C is low, there is much regularization. When C is high, there is low regularization.
epsilon: is used to control the width of the street.
from sklearn.model_selection import RandomizedSearchCV
params = {'gamma':[0.0001, 0.1],'C':[1,1000], 'epsilon':[0,0.5], 'degree':[2,5]}
rnd_search = RandomizedSearchCV(SVR(), params, n_iter=10, verbose=2, cv=3, random_state=42)
rnd_search.fit(training_data_preprocessed, training_labels)
Fitting 3 folds for each of 10 candidates, totalling 30 fits
[CV] END .............C=1, degree=2, epsilon=0, gamma=0.0001; total time= 17.0s
[CV] END .............C=1, degree=2, epsilon=0, gamma=0.0001; total time= 16.2s
[CV] END .............C=1, degree=2, epsilon=0, gamma=0.0001; total time= 15.4s
[CV] END ................C=1, degree=2, epsilon=0, gamma=0.1; total time= 15.1s
[CV] END ................C=1, degree=2, epsilon=0, gamma=0.1; total time= 15.2s
[CV] END ................C=1, degree=2, epsilon=0, gamma=0.1; total time= 15.1s
[CV] END ................C=1, degree=5, epsilon=0, gamma=0.1; total time= 15.4s
[CV] END ................C=1, degree=5, epsilon=0, gamma=0.1; total time= 16.7s
[CV] END ................C=1, degree=5, epsilon=0, gamma=0.1; total time= 14.8s
[CV] END ........C=1000, degree=5, epsilon=0.5, gamma=0.0001; total time= 15.1s
[CV] END ........C=1000, degree=5, epsilon=0.5, gamma=0.0001; total time= 15.3s
[CV] END ........C=1000, degree=5, epsilon=0.5, gamma=0.0001; total time= 16.9s
[CV] END .............C=1000, degree=5, epsilon=0, gamma=0.1; total time= 17.8s
[CV] END .............C=1000, degree=5, epsilon=0, gamma=0.1; total time= 20.4s
[CV] END .............C=1000, degree=5, epsilon=0, gamma=0.1; total time= 18.1s
[CV] END ...........C=1000, degree=2, epsilon=0.5, gamma=0.1; total time= 20.8s
[CV] END ...........C=1000, degree=2, epsilon=0.5, gamma=0.1; total time= 23.3s
[CV] END ...........C=1000, degree=2, epsilon=0.5, gamma=0.1; total time= 22.2s
[CV] END ..........C=1000, degree=2, epsilon=0, gamma=0.0001; total time= 20.6s
[CV] END ..........C=1000, degree=2, epsilon=0, gamma=0.0001; total time= 20.1s
[CV] END ..........C=1000, degree=2, epsilon=0, gamma=0.0001; total time= 20.3s
[CV] END .............C=1000, degree=2, epsilon=0, gamma=0.1; total time= 19.0s
[CV] END .............C=1000, degree=2, epsilon=0, gamma=0.1; total time= 18.9s
[CV] END .............C=1000, degree=2, epsilon=0, gamma=0.1; total time= 19.7s
[CV] END ...........C=1, degree=2, epsilon=0.5, gamma=0.0001; total time= 23.5s
[CV] END ...........C=1, degree=2, epsilon=0.5, gamma=0.0001; total time= 27.7s
[CV] END ...........C=1, degree=2, epsilon=0.5, gamma=0.0001; total time= 17.9s
[CV] END ...........C=1000, degree=5, epsilon=0.5, gamma=0.1; total time= 16.7s
[CV] END ...........C=1000, degree=5, epsilon=0.5, gamma=0.1; total time= 16.7s
[CV] END ...........C=1000, degree=5, epsilon=0.5, gamma=0.1; total time= 15.2s
RandomizedSearchCV(cv=3, estimator=SVR(),
param_distributions={'C': [1, 1000], 'degree': [2, 5],
'epsilon': [0, 0.5],
'gamma': [0.0001, 0.1]},
random_state=42, verbose=2)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomizedSearchCV(cv=3, estimator=SVR(),
param_distributions={'C': [1, 1000], 'degree': [2, 5],
'epsilon': [0, 0.5],
'gamma': [0.0001, 0.1]},
random_state=42, verbose=2)SVR()
SVR()
We can now find the best parameters
rnd_search.best_params_
{'gamma': 0.1, 'epsilon': 0, 'degree': 5, 'C': 1000}
svr_rnd = rnd_search.best_estimator_.fit(training_data_preprocessed, training_labels)
Now, let’s evaluate it on the training set
predictions = svr_rnd.predict(training_data_preprocessed)
mse = mean_squared_error(training_labels, predictions)
rmse = np.sqrt(mse)
rmse
68684.15262765526
Wow, this is much bettter. We were able to improve reduce the root mean squarred error from $117,513 to $68,684.
Now, we can evaluate the model on the test set. We will have to transform it the same way we transformed the training set for the prediction to be possible.
test_input_data = test_data.drop('median_house_value', axis=1)
test_labels = test_data['median_house_value']
test_preprocessed = final_pipe.transform(test_input_data)
test_pred = svr_rnd.predict(test_preprocessed)
test_mse = mean_squared_error(test_labels,test_pred)
test_rmse = np.sqrt(test_mse)
test_rmse
68478.07737338323
That’s is not really bad.
This is the end of the notebook. It was a practical introduction to using Support Vector Machines for regression. In the next lab, we will take a further step, where we will do classification with SVM.
42.66.9. Acknowledgments#
Thanks to Jake VanderPlas for creating the open-source course Python Data Science Handbook . It inspires the majority of the content in this chapter.


